
Happy New Year 2025! Boas has accepted an offer from the University of Bonn (Professorship for Molecular Plant Sciences). The PuckerLab will therefore soon be relocating to Bonn, where we will continue our research on various biosynthesis pathways and their evolution. We are looking forward to expanding the team, new collaborations, and many exciting projects. More details will be shared on the new PuckerLab website soon.
Ronja Friedhoff was awarded the "Bürgerpreis 2024" for her exceptional activities in research and her volunteer engagement. Among her significant achievements are the sequencing of numerous plant genomes, an invited lecture at the ONT conference 'LondonCalling 2024,' and a very successful participation in iGEM 2023 ('Best Diagnostics Project'). During her Bachelor's studies, Ronja already achieved significant research results during a stay abroad in Canada and published them in PLOS ONE. In addition, she is actively involved in student affairs within the department and mentors the SynBio2024 team. Some selected achievements were acknowledged by Boas in a laudation. Ronja is now pursuing a career in virology.
Boas visited the Leibniz Institute DSMZ (German Collection of Microorganisms and Cell Cultures). He learned about current research projects and discussed potential collaborations. Afterwards, he gave a talk titled 'Unlocking Plant Genomes.'

Boas highlighted in his lecture as part of the Studium Generale how genomic research can contribute to sustainable food production. As examples, he presented nematode-tolerant sugar beets and virus-resistant cassava plants. Genomics helps to understand the genetic basis for such desirable traits.
Boas presented his career and the research of the working group at Burg Warberg to the new biology students. The Nanopore sequencing and spectacular plants, such as Victoria cruziana, were of particular interest. The students were invited to sign up for a lab tour if they were interested.
Melina has visited WYMM in Bonn to learn about latest developments regarding ONT sequencing.
Researchers gave presentations covering topics ranging from human genomics for rare diseases to sequencing in cancer research. A particular focus was on identifying and analyzing structural variants and methylation, enabled by the Nanopore technology. Alongside the informative presentations, multiple networking sessions allowed participants to engage with professionals and Nanopore experts, fostering valuable connections.

Nancy, Maria, Chiara, Jakob, Milan and Boas are attending the International Conference of the German Society for Plant Sciences 2024 in Halle. They present the ongoing research projects of our group and look forward to inspiring discussions. The team is open to new collaborations.
Please find an overview of our posters below:
Nancy: P035 "Comparative genomics reveals a conserved biosynthetic gene cluster in withanolide-producing Solanaceae species"
Maria: P367 "Evolutionary “Hotspots” in Flavonoid Biosynthesis"
Jakob: P061 "Genome Sequence of the ornamental plant Digitalis purpurea reveals the molecular basis of flower traits"
Chiara: P324 "The Science of Blue Flowers: Phylogenetic, Genomic and Transcriptomic Analysis of Blue Flowering Plants"
Milan: P377 "Uncovering the Evolution of Anthocyanin-Related Glutathione-S-Transferases"
The full programme of the conference is available here.

Our SynBio2024 team was featured in the latest issue of BIOspektrum. The article describes the project BlueBloom. Here you can find the article.

Within a week, doctoral students from various research institutions in Braunschweig (TU Braunschweig, JKI, HZI) acquired fundamental skills in genomics, transcriptomics, and applied bioinformatics. Additionally, they learned strategies on how to independently install and use new bioinformatics tools in the future. Further details are described in the TU Magazine article.
This week, we are hosting a group of 15 students from the Universidad Nacional Agraria La Molina at TU Braunschweig. On Tuesday, the students received information about studying in Germany from our international house. On Wednesday, scientific presentations by young scientists in Braunschweig highlighted some research directions. On Thursday, we visited the Botanical Garden and the Institute of Pharmaceutical Biology with its garden of medicinal plants. A barbecue concluded the day and offered plenty of time for questions and discussions. On Friday, our guests were guided through the biocenter and BRICS to learn about large analytical instruments. Afterwards, we visited the JKI campus with tours through the cassava farm and the hop garden. On Saturday, our guests also gave presentations about their research projects that sparked discussions. The visit concluded with a final summary presentation by Boas, and the students continued their journey by visiting the university in Hohenheim. More details are described in this article on the TU blog.
Ronja Friedhoff reports in the TU Magazine article "Successful research while still studying" about her experiences with ONT sequencing. Using various genome sequencing projects on medically relevant plants as examples, she presents the potential of genomic analyses. Since Ronja is particularly interested in viruses, she has investigated biosynthetic pathways of potentially antiviral substances from plants. There is a video recording of the lecture on the LondonCalling2024 platform by ONT.

Nele, Samuel, and Boas used the TalentCampus to learn about crop science at KWS.

Kiersten (Arizona State University) has begun her DAAD-funded research stay at TU Braunschweig by harvesting cacao leaves. She aims to extract DNA from these leaves for nanopore sequencing. Sequencing will decode the plant's genome. By comparing the genomes of different cacao cultivars, the genetic basis for differences in the properties of these cultivars will be identified. Kiersten is being supervised by Maria, who offered this project as part of the RISE program.
Ronja and Katharina are presenting our research at the LondonCalling2024 conference organized by ONT. Ronja will introduce various genome sequencing projects on medically relevant plants and explain how these form the basis for elucidating biosynthetic pathways. One example is Digitalis purpurea, which is also known for fascinating differences in flower color (Wolff, Friedhoff, Horz, Pucker, 2024). Katharina will present our experiences from the "Data Literacy in Plant Genomics" courses. Details on this have already been described in a publication (Wolff et al., 2023).
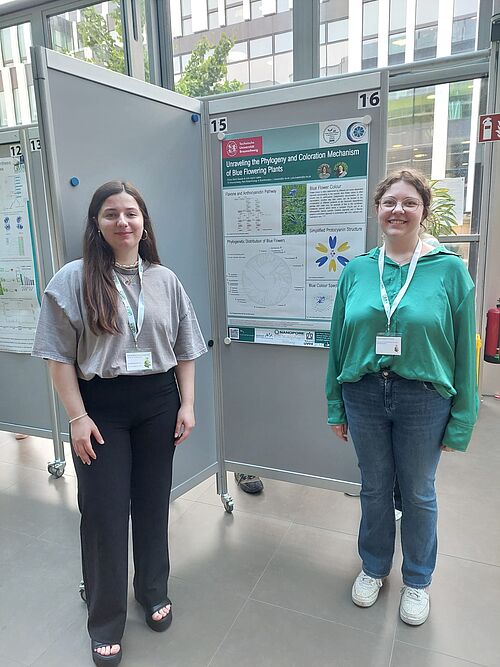
Chiara Dassow and Julia Lüpkes presented the SynBio2024 project at the Mendel Early Career Symposium 2024 in Vienna. Further details about this project are described on the #SynBio2024 website.

Boas presented the group's research with a lecture on the topic "Exploring the evolution of biosynthesis networks with big data" at the IPB Sympoium 2024 in the premises of the Leopoldina in Halle. Among other things, work on the competition between DFR and FLS (Choudhary & Pucker, 2024), a systematic analysis of the independent loss of anthocyanin biosynthesis in different plant species (Recinos & Pucker, 2024), KIPEs3 (Rempel et al., 2023) and various genome sequencing projects were presented (Wolff et al., 2024).

During the seminar "Meet The BRICS," Boas presented the research of the group, and Nancy specifically showcased some research on flavonoid biosynthesis. This focused particularly on KIPEs3 and the opportunity it creates for analyzing large datasets. As an example, the analysis of the substrate specificity of DFR and FLS was presented.

Ronja, Julia, and Boas visited the Braunschweiger Bürgerstiftung (Braunschweig Citizens' Foundation) to report on the successes of iGEM 2023 and discuss plans for SynBio2024. Ronja provided a detailed presentation on how the 2023 iGEM team from TU Braunschweig received awards at the iGEM Giant Jamboree with the project 'Li+onSwitch.' Boas explained the reasons for opting out of future iGEM participation and outlined plans for an alternative project. We discussed our plans for science communication, especially in conveying synthetic biology content to high school students. The foundation has already supported our teams in 2022 and 2023, and we look forward to a fruitful collaboration in 2024.

Nancy presented the results of her Master thesis at MBP2024 with a poster. More details about this topic are available through the corresponding preprint describing the competition of DFR and FLS.

Digitalis purpurea (Foxglove) is known for an intense purple/magenta flower color. However, some years ago, we observed white flowers. Immediately, the question arose regarding the genetic cause for this difference. The color is very likely due to anthocyanins. Therefore, we suspected a blockade in the anthocyanin biosynthetic pathway. But which gene could have been disrupted by a mutation in plants with white flowers? This mystery has now been solved by Katharina, Ronja, and Jakob through sequencing the genome of D. purpurea. A comparison of the genomes of plants with purple/magenta flowers against the genomes of plants with white flowers revealed a significant difference: white plants carry a large insertion in a gene encoding an anthocyanidin synthase. The loss of this gene could block the biosynthesis of anthocyanins. Further details of our analyses are described in a preprint on bioRxiv.

The Christmas party last Friday was the festive highlight of a very successful week. In the lovingly decorated social room, we enjoyed a variety of delicious treats and spent a lovely evening. There was even a Christmas tree and a visit from Santa Claus (Benni), who distributed the gifts for our 'Schrottwichteln' (a fun gift exchange).

Corinna Thoben was awarded a prize for outstanding achievements during her bachelor's studies. Key factors for this award were her excellent academic performance and her social commitment, particularly in conveying basic biological principles to students. Additional details about the award ceremony and backgrounds are provided in an article in the TU Magazin.

On December 1st, we visited the Christmas market in the historical city center of Braunschweig. There was mulled wine, bratwurst, and many other delicious treats.

Ronja Friedhoff and Susanna Pape gave a final presentation about the iGEM2023 project Li+onSwitch at the BRICS. Interested relatives, students, and faculty had the opportunity to learn details about the project and to ask questions. This event celebrated the team's excellent achievements: Best Diagnostics Project, Top 10 Teams, and a Gold Medal. We wish the entire team much success in their future projects!

Today, Samar Sheat presented her cassava research in the AG Winter (DSMZ) to a group of students at TU Braunschweig. Cassava is an important crop grown on several continents. Samar is currently looking for resistance genes against viral infections that are currently destroying yields. Support is needed for this research. Interested students are welcome to get in touch.
The huge success of the iGEM2023 team of TU Braunschweig is featured in various media (some in German):

Boas gave at talk with the title "Investigating plant specialized metabolism through genomics and applied bioinformatics" in the joined colloquium of DSMZ and JKI.

Our iGEM2023 team won the 'Best Diagnostics' award and a gold medal. Additionally, the team was nominated for the 'Best Composite Part' award. The project Li+onSwitch aiming at the quantification of lithium levels in blood samples convinced the judges at the Grand Jamboree in Paris. A detailed description of the project is available in the team wiki. Moreover, there are several videos about the team's project: project promotion video.
Interested in Synthetic Biology? We are recruiting a new team for an exciting project: SynBio @ TU Braunschweig.

On 3rd November 2023, biology and biotechnology students at TU Braunschweig received their graduation certificates. Huge congratulations to Melina, Sina and Benni for earning a Bachelor of Science degree in biology!

Boas gave a talk with the title "Pioneer in Genomes" in the seminar "Innovations from biotechnological research - trends and techniques that shape the future" at Bielefeld University and discussed carrier options with students.

We are offering some information events for students interested in synthetic biology. The aim is to enable a student team to work on a synbio project during the next months. Students who are interested, but cannot participate, are asked to send an email.

The Ecki Wohlgehagen Foundation has supported the purchase of a new thermocycler for molecular biology experiments at the TU Braunschweig. With this device, reaction mixtures can be incubated under a variety of precisely controlled temperatures. This is important for controlling molecular reactions and the activity of enzymes. The spectrum of possible applications ranges from routine methods such as PCR for amplification of DNA molecules to sample preparation for modern nanopore sequencing.
The board of trustees of the Ecki Wohlgehagen Foundation, represented by Mr. Wohlgehagen and Mr. Smyrek, visited the AG Pucker laboratories and had the new device and various possible applications explained to them. “Through the support of the Ecki Wohlgehagen Foundation, we can provide students with practical experience in the use of state-of-the-art laboratory methods and thus ensure the high level of life science education at the TU Braunschweig,” says Prof. Dr. Boaz Pucker.
Life science students use this device in courses on plant molecular biology and genomics. The iGEM team at TU Braunschweig also benefits from this new acquisition because additional capacity for DNA replication has been created. “The thermal cycler was very helpful for our iGEM project because we had to carry out a particularly large number of clonings and PCRs,” says Corinna Thoben from the iGEM team at TU-Braunschweig 2023. “DNA amplification and the combination of different pieces of DNA via PCR provides "The basis for many applications in synthetic biology," adds Ronja Friedhoff, who is also a member of the iGEM team. In the future, other students will work on their synthetic biology projects with this device. Among other things, projects are planned to produce glowing plants to create the targeted introduction of the necessary genes. Students should use this thermal cycler to prepare their samples for nanopore sequencing in courses on plant genome sequencing.
The Ecki Wohlgehagen Foundation is managed in trust by the Braunschweig Community Foundation and supports numerous educational measures.

Boas gave a talk with the title "Exploring biosynthesis pathways in fungi and plants" as part of the workshop "Microbial Interactions in the Phytosphere" and discussed future projects with researchers of different institutions.

On 10th October, Boas introduced himself and the research of the group to Faculty 2 during an inaugural lecture. Title of the lecture was "Big data-driven discoveries in the specialized plant metabolism". Selected research projects of the group were presented around three methods (comparative genomics, coexpression, applied bioinformatics). Afterwards, there was plenty of time for questions and discussions at the buffet.
Jen and Sarah share their internship experiences in the TU Magazine.
This week, Michael and Jen left our group to continue their studies at their home universities. Michael is now working at Uni Marburg to earn his doctoral degree. Jen is back at UC San Diego to complete her undergraduate studies. Michael performed an amazing coexpression analysis on Arabidopsis thaliana data to identify novel genes in the anthocyanin biosynthesis. Many thanks for the excellent help in setting up the lab and also supporting the iGEM teams in 2022 and 2023! Jen studied cacao genomes to understand their specific properties. All the best for the future!

Boas presented some research of the group in his talk about comparative genomics at the section meeting biodiversity & evolution 2023 in Gießen. It was a successful meeting with many fruitful discussions, which might lead to future collaborations. The photo shows a figure from a study about the evolution of the Apiaceae FNS I (Pucker & Iorizzo, 2023).

Lena Fürstenberg examined the anthocyanin biosynthesis in the stinging nettle (Urtica dioica) for her bachelor's thesis. The medicinal plant of the year 2022 is renowned for producing various flavonoids, including anthocyanins. These compounds are formed in response to stress and accumulated within the plant, resulting in an intense red coloring of the leaves. The findings of this study were presented and discussed with the research group.
Rheum nobile grows in the Himalayas and is famous for its adaptation to the extreme conditions at high altitudes. The plant has an outer layer of leaves that serve for insulation and protection against UV radiation. This helps to increase the temperature inside the plant and safeguard the vital parts of the plant from the harmful effects of UV radiation. The genome sequence of Rheum nobile has now been published by Feng, Pucker, Kuang et al., 2023, providing an important foundation for understanding the biology of this species.
Our new BioInfTool server is online and allows the use of various bioinformatics tools without any installation. Through a web browser, genes involved in flavonoid biosynthesis can be identified in new plant species, or transcription factor families can be investigated. We welcome feedback and suggestions for additional tools that we should include in the future. Many thanks to Andreas Rempel and de.NBI for their excellent support!

Boas visited the research group Biochemistry of Plant Specialised Metabolites (University of Hannover) and enjoyed numerous discussions about biotechnology and specialized metabolism in plants. As part of a seminar, various projects for omics-based elucidation of biosynthetic pathways were presented. Many thanks for the invitation and the amazing time in Hannover!

Out iGEM2023 team was featured in TU Magazin. The article presented the project idea and shared the experiences gained through a recent presentation in the BRICS by Ronja and Susanna.

We have performed direct RNA sequencing using our MinION. Melina extracted RNAs from flower samples of an aquatic plant. Melina and Benni prepared these RNAs for sequencing by creating complementary cDNA and attaching adaptors. After quality control, sequencing was performed on an R9 flow cell using one of our MinIONs. The length distribution shows an average read length of several hundred nucleotides, as many mRNAs are quite short. These data will provide an important foundation for gene identification. Many exciting experiments using this technology are planned!
As part of her Bachelor thesis, Corinna Thoben developed a tool for the automatic identification and annotation of bHLH transcription factors in the genome sequences of plants. A corresponding preprint was just published on bioRxiv: Automatic annotation of the bHLH gene family in plants. The bHLH_annotator is freely available via GitHub.

Today, the iGEM team 2023 of TU Braunschweig presented the plan for their project Li+onSwitch in a public talk in the BRICS. All interested members of TU Braunschweig and other research institutes in Braunschweig participated. Ronja and Susanna gave an excellent talk. Additional updates will be shared through the team's website.
All teaching materials of our "Data Literacy in Genome Research" course are now available through Twillo. The development of this course is funded by the Stiftung Innovation in der Hochschullehre. All files are available on github, but we hope for an increased visibility through Twillo. Additional information about this teaching innovation are available on the DaLiP website.

Our iGEM team TU Braunschweig 2023 was interviewed by the local newspaper. Now, you can find the content of that interview in the published newspaper article (in German).

Boas visited the research group Systematics, Biodiversity and Evolution of Plants (LMU Munich) and enjoyed numerous discussions about the evolution of biosynthesis pathways. The invited talk covered our recent projects about the evolution of biosynthesis pathways with a focus on genomics. Plants in the greenhouses of the Botanical Garden Munich-Nymphenberg are impressive. Many thanks for the invitation and the great time in Munich!

Boas visited the research group Physiology of Yield Stability (University of Hohenheim) and enjoyed inspiring discussions about the biosynthesis of specialized metabolites. The invited talk covered the recent insights into the mutual exclusion of anthocyanins and betalains in the Caryophyllales. Many thanks for the invitation and the excellent organisation!
The experiences and success of the iGEM team TU Braunschweig 2022 were summarized in the newsletter of the Braunschweiger Hochschulbunds (BHB) (in German) by Christian Köcher. Please find details about the 'LionDetect' project there.
Due to the nice weather we decided to have a last barbecue in 2022. There is always something to celebrate and this is a great opportunity for exchange in the team.
For over 50 years, researchers have investigated the red, yellow, and blue pigments of plants in the Caryophyllales. Only plants in this order can have betalains, while most other land plants are pigmented by anthocyanins. However, the betalain-pigmented Caryophyllales plants do not accumulate anthocyanins. Recent studies by Fan et al., 2020 (now retracted) and Zhou et al., 2020 claimed that anthocyanins would contribute to the pigmentation of red pitayas. We previously addressed issues in the Fan et al., 2020 paper in our response (Pucker et al., 2022). Now, the paper by Zhou et al., 2020 was addressed by two correspondence articles just published in BMC Genomics. Both refute the contribution of anthocyanins to pigmentation in pitaya:
Pucker, B., Brockington, S.F. The evidence for anthocyanins in the betalain-pigmented genus Hylocereus is weak. BMC Genomics 23, 739 (2022). https://doi.org/10.1186/s12864-022-08947-1
Khan, M.I. The reported colour formation mechanism in pitaya fruit through co-accumulation of anthocyanins and betalains is inconsistent and fails to establish the co-accumulation. BMC Genomics 23, 740 (2022). https://doi.org/10.1186/s12864-022-08957-z
If you are interested in this topic, please have a look at these recent publications: Timoneda et al., 2019, Sheehan et al., 2020, and Pucker et al., 2022.
Shakunthala shares her experiences in Germany in a recent blog post. A particular focus is on her research experiences in our group at TU Braunschweig.
Maria offers a project with the title
Unwrapping the wonders of chocolate: from candy to functional food
for international students as part of the RISE program of the DAAD. Please contact Maria directly if you are interested in additional details. Would you like to apply? You can do this through the DAAD portal. Here is the project code: Braunschweig_BI_CH_4798.
Here is the blog post about the future of long read sequencing in plant genomics: "From finding flavonoids to the ‘dark matter’ of T-DNA insertions — investigating plant genomes with nanopore sequencing".
Boas was invited to give a talk about applications of long read sequencing in plant genomics as part of the APBioNET seminar series.
Life science students learnt the basis of Python3 and completed several application exercises. Slides and materials are available via github. Students can expressed there interest in taking this course in the future can send an email to Boas Pucker.
Melina and Benni (s. photo) are excited about the flower color change of Victoria spec. They collected samples during the summer of 2022 and would like to investigate these in detail to understand the molecular mechanisms. Therefore, they are collecting support through a crowd funding project at betterplace.
Many thanks to de.NBI for excellent support! Our research would not be possible without de.NBI. The details are described in a testimonial on the de.NBI website.
Successful (international) collaborations resulted in three new publications:
A proposal for "Freiraum 2022" received very positive evaluations and funding. As of September 2022, we are preparing several courses around 'plant genomics' and 'data literacy'. More details are available here (in German).
Boas Pucker gave a presentation at International Conference of the German Society for Plant Sciences 2022 in Bonn. Topic of this talk were molecular mechanisms that explain mutual exclusion of anthocyanins and betalains in the Caryophyllales. This involved KIPEs which was important for the identification of anthocyanin biosynthesis genes.
We released an updated version of KIPEs on our web server. The changes between KIPEs3 and the original KIPEs version are described in a prepint:
We started the sequencing of two plant genomes. The picture shows the state of nanopores during the sequencing process. Green indicates that the nanopores are active and sequence DNA strands. First analysis of the datasets are promising. More details about this innovative sequencing technology are available in our recent review article.
A recent article in TU Magazin reports about the iGEM team TU_Braunschweig 2022, which is working on the laboratories of our group. The endless possibilities of an iGEM participation are highlighted in this article.
The iGEM team TU_Braunschweig 2022 is now officially registered for the competition. iGEM is a synthetic biology competition for student teams from around the world. Teams of TU Braunschweig participated very successfully in 2013 and 2014.
Our bioinformatic tools (MYB_annotator & KIPEs) are now available through a web server. This web server enables researchers without any bioinformatic skills to benefit from these tools. Files with sequences are uploaded to start the analysis. Once the analysis is completed, a compressed archive of all result files will be offered for download. Currently, the choices of parameters are restricted. The addition of other tools is in progress.
We wrote a review article about the development and current state of long read sequencing technologies with focus on plant genomics. This review covers the sequencing technologies provided by ONT and PacBio as well as applications of these technologies. Future challenges of plant genomics are presented.
Our publication "Comparison of Read Mapping and Variant Calling Tools for the Analysis of Plant NGS Data" was selected by the Editor as one of the promising and important studies in the field. We compared different bioinformatic tools for the alignment of reads against a reference sequence and the following identification of sequence differences.
We are currently informing students at TU Braunschweig about the opportunity to participate in iGEM, the international competition for synthetic biology. More details are available here: iGEM (in German).
An interview with Boas Pucker was published under the title "In Search of Drug Candidates in the Plant Genome" in the TU Magazin.
In October, the results of three projects were published as preprints on bioRxiv:
Boas Pucker was accepted as member of the Braunschweig Integrated Centre of Systems Biology (BRICS) on the 11th October 2021.
Since the 1st October 2021, the group is located at the TU Braunschweig in the Institute of Plant Biology (Mendelssohnstrasse 4). Details about research and resulting publications are available on the dedicated pages.